top of page
ENZYME ACTIVITY MODULATORS
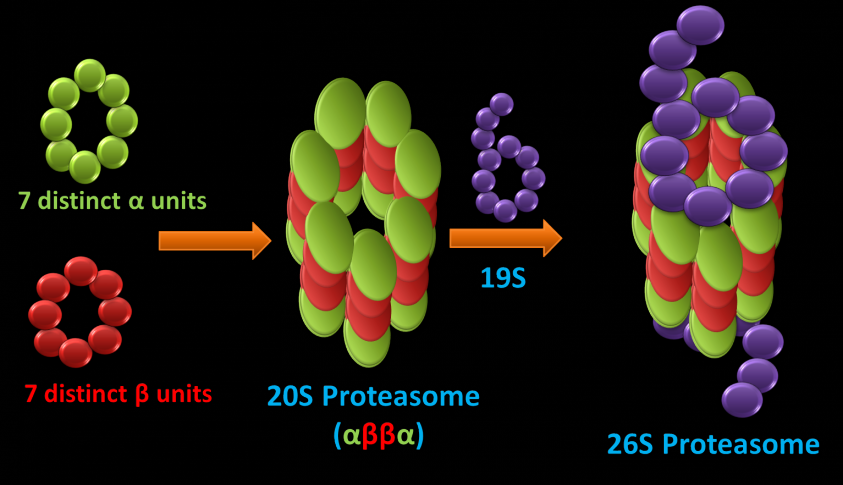
Since the discovery of lysosomes, it was believed that this organelle degrades cellular proteins. However, several independent researchers showed strong evidence for non-lysosomal intracellular protein degradation, but this process's mechanisms were unclear until proteasome discovery. The ubiquitin-proteasome pathway is a major avenue for the degradation of proteins in the cytosol and nucleus of eukaryotic cells. Proteins are marked for degradation via the attachment of a ubiquitin group whereby the proteasome recognizes, unfold, and digests these proteins. The 26S proteasome is a cylindrical shaped proteolytic complex (Figure 1 (left)) that incorporates the ATP-dependent 19S caps, which recognize the proteins marked for degradation and unfold substrates in turn fed to the 20S proteasome, where proteolysis occurs. 20S core particle is a four stacked heptameric ring composed of a and B units (a 7, B 7, B 7, a 7). subunits guard the entrance to the active site by allowing access to only unfolded proteins.
The catalytic activities are confined to the B subunits responsible for mediating the enzymatic activity of the proteasome. Three of the B-subunits in the B-ring are catalytically active, and they have an N-terminal threonine (side-chain hydroxyl group activated by the N-terminal amine moiety) that acts as the proteolytic nucleophile (Figure 1 (right)). The proteasome has at least three distinct peptidase activities and, by comparison with substrate specificities of known proteases, they are designated as chymotrypsin-like (B5-subunit), trypsin-like (B2-subunit), and caspase-like activities (B1- subunit).
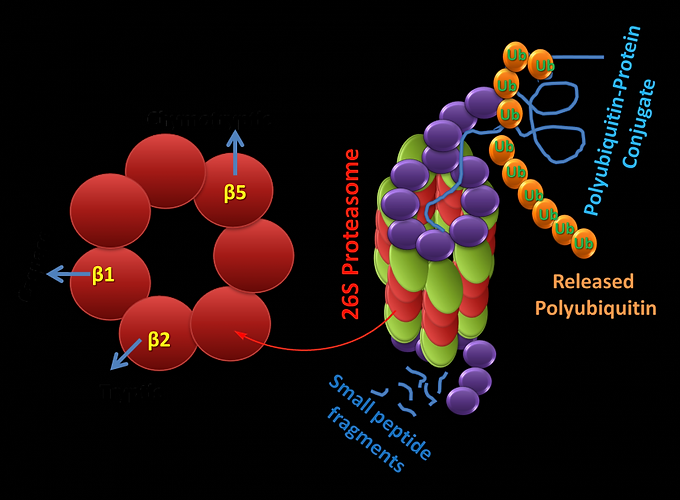
Figure: Protolytically active B units (left). Active sites located within the 20S core particles hydrolyze these unfolded polypeptides into small peptides and amino acids (Right). Proteins synthesized in the cell (direct presentation) or are released from endosomes (cross-presentation) are polyubiquitylated in the cytoplasm and degraded by hybrid proteasomes consisting of the 20S proteasome core, the 19S regulator. The peptides produced are either of the ideal lengths for binding to MHC class I molecules or are amino-terminally extended precursors that can be further cleaved by aminopeptidases in the cytoplasm.
It removes abnormal proteins that may be misfolded, aged, or damaged by oxidation. It also regulates the half-life of the short-lived regulatory proteins such as cyclins involved in controlling the cell cycle and transcription regulators such as p53 and NF-kB. Through its destruction function, the proteasome is thus involved in various cellular processes such as protein quality control, cell cycle, antigen presentation, apoptosis, and cell signaling. Proteasomes are also found in several bacterial species of the order Actinomycetales. Numerous studies demonstrated that mycobacterium tuberculosis (Mtb), an Actinomycete pathogenic to humans, involves proteasome function to root infections. Thus, understanding the proteasome function in Mtb would help get new insight into how the host combats infections. Thus, proteasome becomes an attractive therapeutic target for controlling various diseases, including cancer.

The active site of the proteasome is nothing but Lewis's basic hydroxyl groups. Obviously, a simple Lewis acid can be employed to control the activity of the proteasome. Our research group has been actively involved in exploiting the Lewis acidity of tricoordinate boron to develop boron-based proteasome inhibitors (BPI) and understand the differences in the catalytic activity of multiple proteolytic sites in both eukaryotic and bacteria proteasomes. Our long-range goals to utilize the acquired knowledge to design novel BPIs to control proteasome activities in specific diseases. We are also actively pursuing the design and development of boronopeptides and their role as antibacterial and diabetic wound healing drug molecules.
bottom of page
